Chapter 2 CCS Database
Unified CCS database aims to be a unified platform to host both literature-reported CCS values and in-silico predicted CCS values for ion mobility - mass spectrometry (IM-MS). It is open-access and downloadable. It contains 3,539 unified CCS values which are summarized from 5,119 experimental CCS records. These experimental CCS values are acquired with variable platform including DTIMS, TWIMS and TIMS etc., and have definitive confidence level. In addition, ~10,000,000 predicted CCS values are provided for ~1,700,000 small molecules from multiple public database to support widespread applications, including metabolomics, lipidomics, drug screening, pesticide screening etc (Table 2.1). For each compound, its compound card contains meta information, complete records and links to other database. Finally, users can search interested compounds’ CCS values with the function of “Browser” and/or “Advance search” in this part.
| No. | Database | Compounds | Coverage | Mirror date | Reference |
|---|---|---|---|---|---|
| 1 | KEGG | 16085 | Metabolites & lipids | 2018-08-02 | Kanehisa and Goto (2000) |
| 2 | HMDB | 113989 | Metabolites & lipids | 2018-06-09 | D. S. Wishart, Feunang, et al. (2017b) |
| 3 | LMSD | 40532 | Metabolites & lipids | 2019-07-11 | Fahy et al. (2008) |
| 4 | MINE | 592175 | Metabolites & lipids | 2018-02-07 | Jeffryes et al. (2015) |
| 5 | DrugBank | 9546 | Drugs & xenobiolics | 2019-04-12 | D. S. Wishart, Feunang, et al. (2017a) |
| 6 | DSSTox | 856919 | Drugs & xenobiolics | 2019-05-06 | M.Grulke et al. (2019) |
| 7 | UNPD | 213188 | Natural products | 2019-06-13 | Gu et al. (2013) |
| 8 | ZhuLab | 1417 | Metabolites & lipids | 2018-09-02 | Zhou et al. (2020) |
2.1 Compound Browser
Compound Browser Function provides a simple and straightforward way to browser the database. There are several browser conditions set in “Browser” part (Figure 2.1).
- Type: It provides the choice of CCS values generated from experiments or prediction.
- Database: this option includes variable databases that cover all compounds in our unified database (Table 2.1). And users can choose specific database(s) for further execution.
- Level: it includes confidence level (See Section 2.2) of compounds in the unified database. It helps to choose compounds in the clearly defined level.
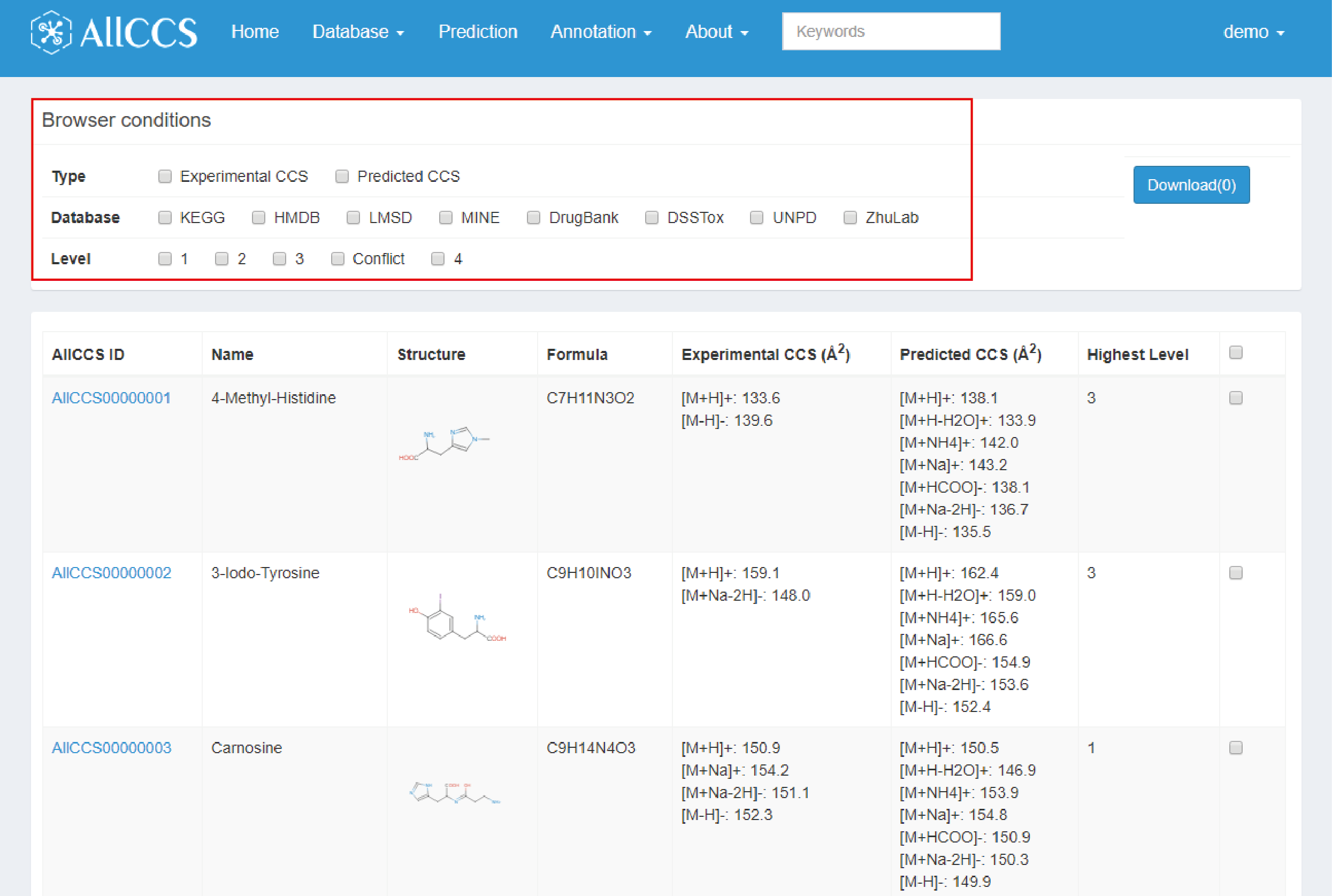
Figure 2.1: Browser conditions
With browser conditions, users can screen out a series of interested compounds. Compounds entries would be displayed according to defined condition. In below text, it contains brief information for each compound.
- AllCCS ID: As described in the section 1.2, users can click the link in the column of AllCCS ID to browse the corresponding compound card (Figure 1.2).
- Name: compound name
- Structure: the image of compound structure
- Formula: chemical formula
- Experimental CCS: The unified CCS value reported in literature (See Section 2.2.3)
- Predicted CCS: The predicted CCS values using machine-learning algorithm. (See Section 2.2.4)
- Highest level: The highest confidence level of CCS values (See Section 2.2.2)
Users can check the interested compounds in the last column. Click the download option, you could download a CSV table containing the information of you checked compounds (Figure 2.2).
Note:
- Download function supports up to 100 items for one time.
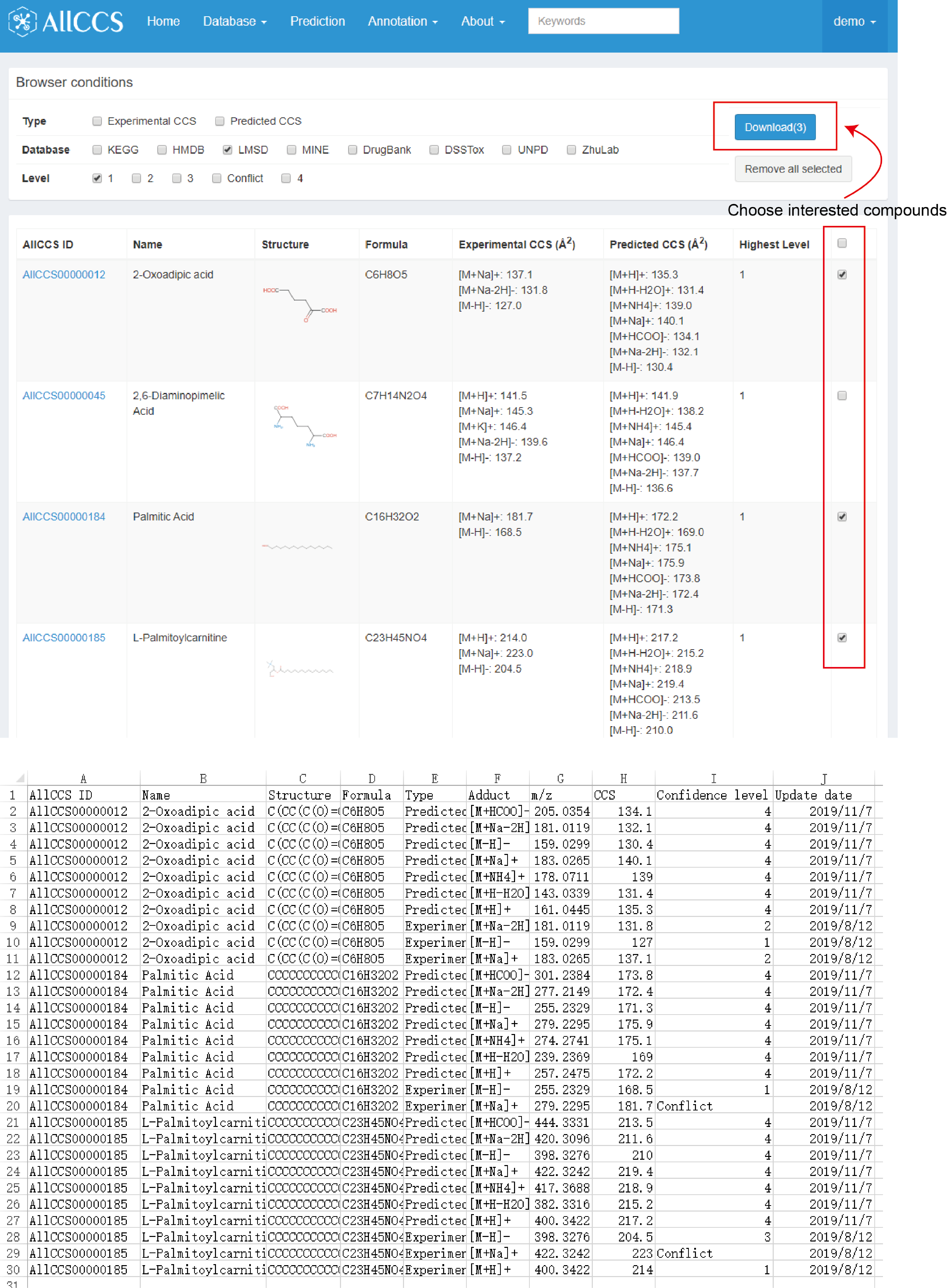
Figure 2.2: Download interested compounds
2.2 Compound Card
In the compound card, it contains detail information of the compound. Next, we would like to explain each parts in the compound card.
2.2.1 Compound information
It contains the basic information of the compound, including ALLCCS ID, name, formula, exact mass, SMILES, InChI, InChIKey, classification and structure in the right panel (Figure 2.3). Here, ClassyFire is used for compounds’ classification (Feunang et al. 2016).
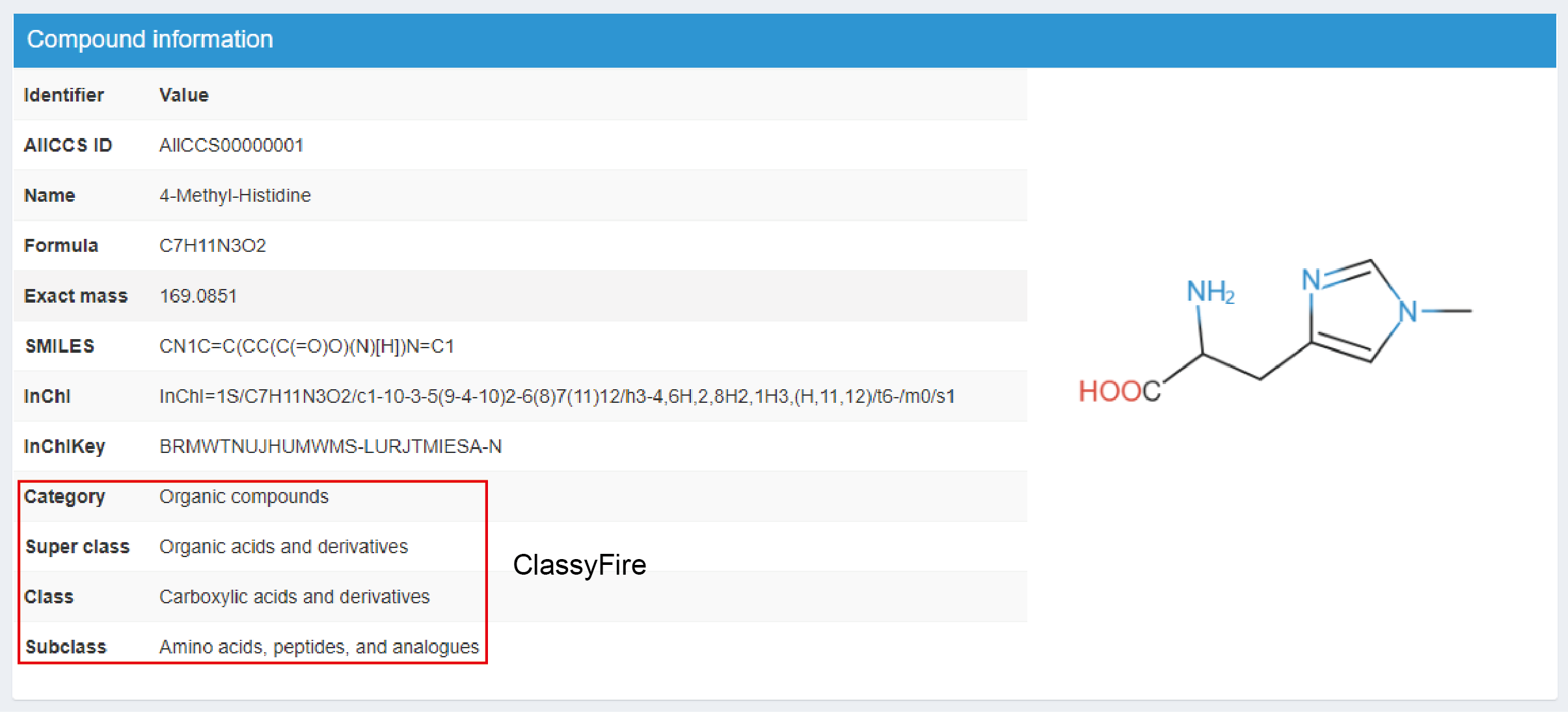
Figure 2.3: Download interested compounds
2.2.2 Unified CCS
This part contains the CCS information of different adduct forms with experimental CCS (if exists) and predicted CCS (Figure 2.4). The CCS reported here is the unified CCS values. We defined unified CCS as the average CCS value with definitive confidence level. The definition of confidence level can find in Table 2.2. CCS values of confidence 1, 2, 3 are all experimental values. The definition of confidence level:
| Confidence level | Platform | Reported labs (N) | Maximum relative error (%) |
|---|---|---|---|
| Level 1 | DTIMS | N≥2 | ≤1% |
| Level 2 | DTIMS/TWIMS/TIMS | N≥2 | ≤3% |
| Level 3 | DTIMS/TWIMS/TIMS | N=1 | — |
| Level 4 | Predicted CCS | — | — |
| Conflict | DTIMS/TWIMS/TIMS | N≥2 | >3% |
- Confidence level 1 represents the CCS value of the specie which is acquired with DTIMS and has been reported at least twice in different labs with the maximum relative error less than 1%.
- Confidence level 2 represents the CCS value of specie which is acquired with DTIMS, TWIMS or TIMS and has been reported at least twice in different labs with the maximum relative error less than 3%.
- Confidence level 3 represents the CCS value of specie is acquired with DTIMS, TWIMS or TIMS and only reported by one lab.
- Confidence level 4 represents the predicted CCS value.
- Conflict means the CCS value of specie which is acquired with DTIMS, TWIMS or TIMS and has been reported at least twice in different labs, but the maximum relative error is more than 3%.
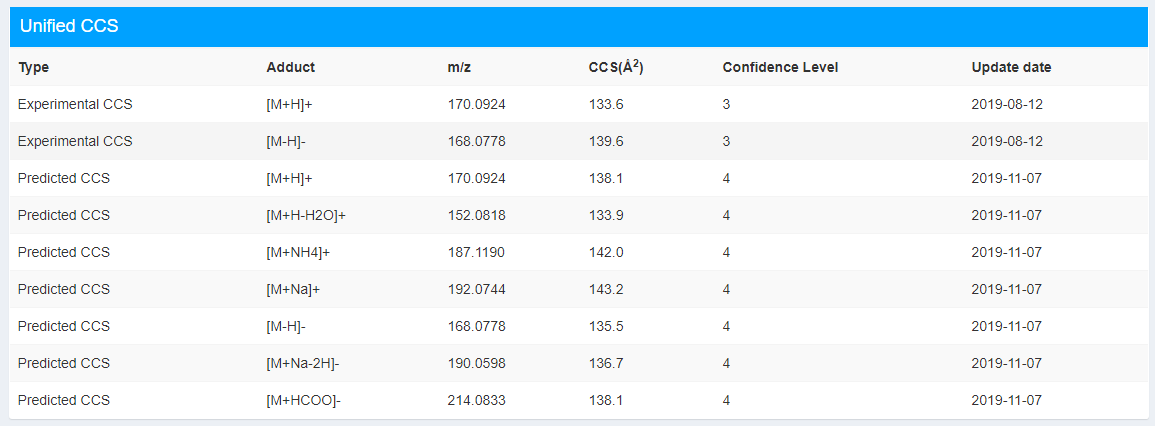
Figure 2.4: Unified CCS
2.2.3 Experimental CCS records
This part records the detailed information of experimental CCS values (Figure 2.5). Compounds that have experimental CCS records contains the basic information of adduct form, m/z, experimental CCS values and charge. Besides, we also provide the information of used instrument platform and the type of ion mobility mass spectrometry. Detail information can be found in Table 2.3. The measured approach is also provided, including single-field, multiple-fields, and empirical method. Corresponding reference literature is listed in the DOI column. If compounds don’t have experimental CCS record, it would have no information in this part.
| Instrument | Type | Vendor |
|---|---|---|
| Agilent 6560 DTIM-QTOF | DTIMS | Agilent |
| Waters Synapt G2-Si HDMS | TWIMS | Waters |
| Waters Synapt G2 HDMS | TWIMS | Waters |
| Waters Vion IMS QTOF | TWIMS | Waters |
| TOFwerk IMS TOF | DTIMS | TOFwerk |
| Bruker timsTOF | TIMS | Bruker |
| Bruker timsTOF Pro | TIMS | Bruker |
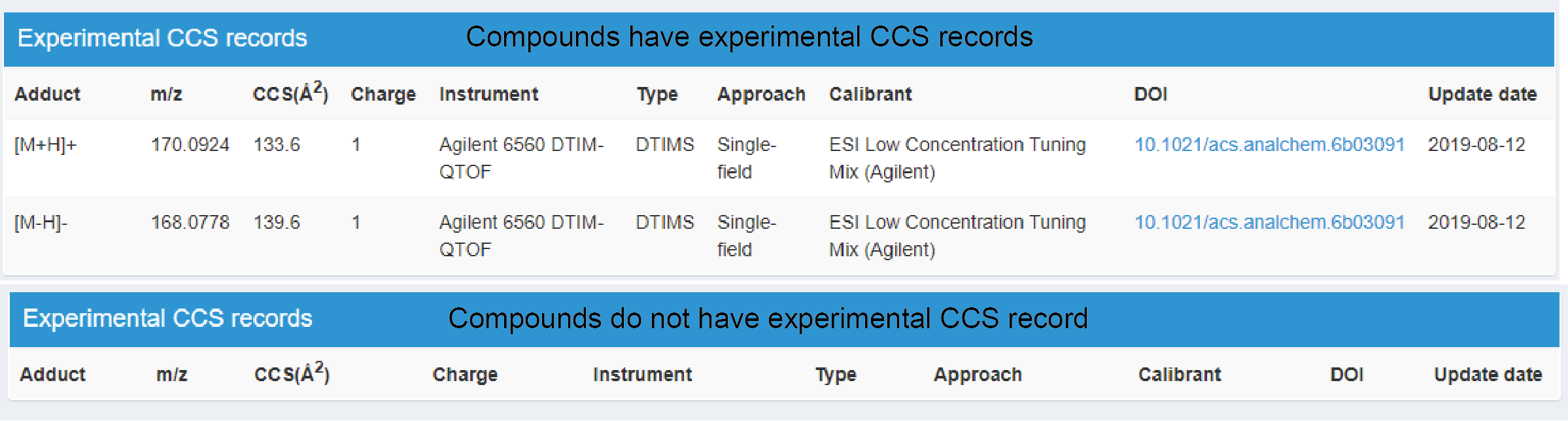
Figure 2.5: Experimental CCS records
2.2.4 Predicted CCS records
This part records the detailed information of predicted CCS values (Figure 2.6). It provides the basic information of adduct forms, m/z, charge, corresponding predicted CCS values using AllCCS_V1 tool, users can reference AllCCS paper for detailed information (Zhou et al. 2020). Here we define representative structure similarity (RSS) to represent the similarity between this compound and the training set.
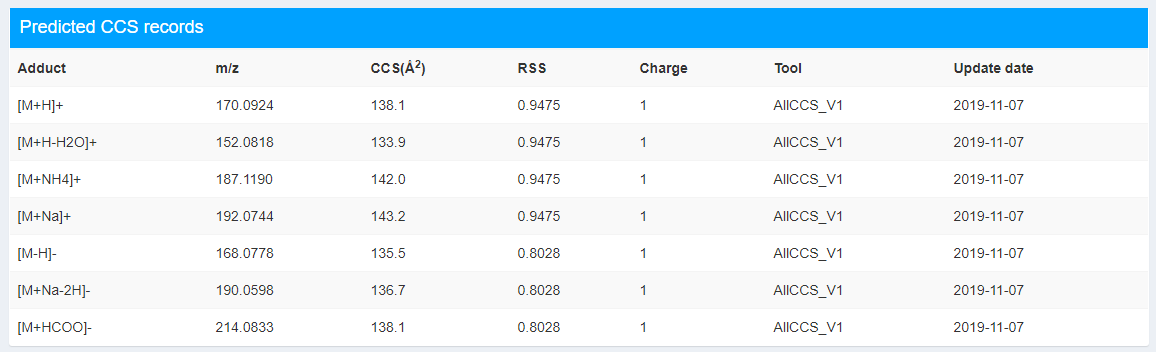
Figure 2.6: Predicted CCS records
2.2.5 Database link
This part provides the link to databases that contain the compound (Figure 2.7).
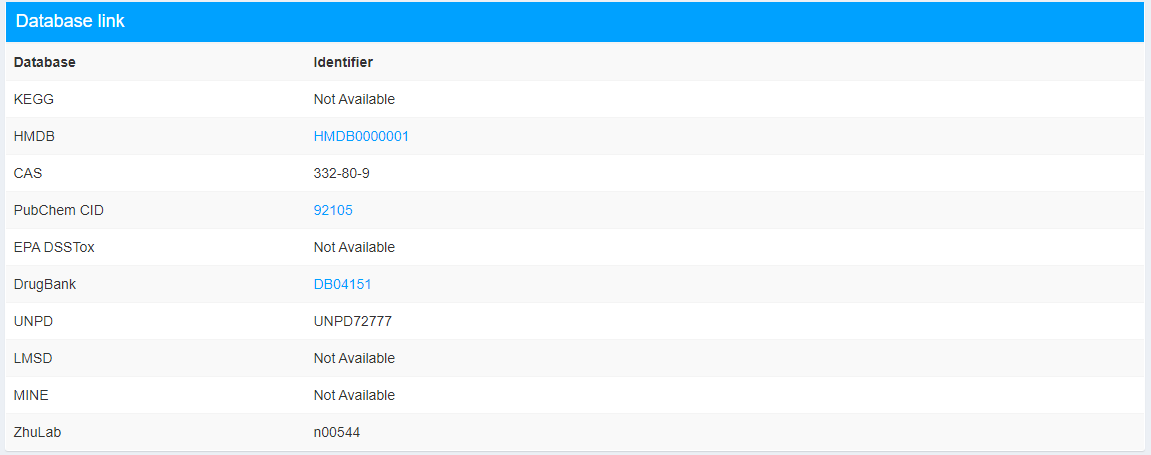
Figure 2.7: Database link
2.3 Advanced Search
For advanced search, there are two modes for users to search the compounds in our unified CCS database, including “single mode” and “batch mode”.
2.3.1 Single mode
As named, users can search for one compounds at one time in single mode. Here, we provide several optional identifiers for users to choose (Figure 2.8), including compound’s name, database ID, formula, SMILES, InChI, InChIKey.
Note:
- If you don’t have confirmed identifier, keep it as null.
- If there are contradictory identifier, it will return no available data.
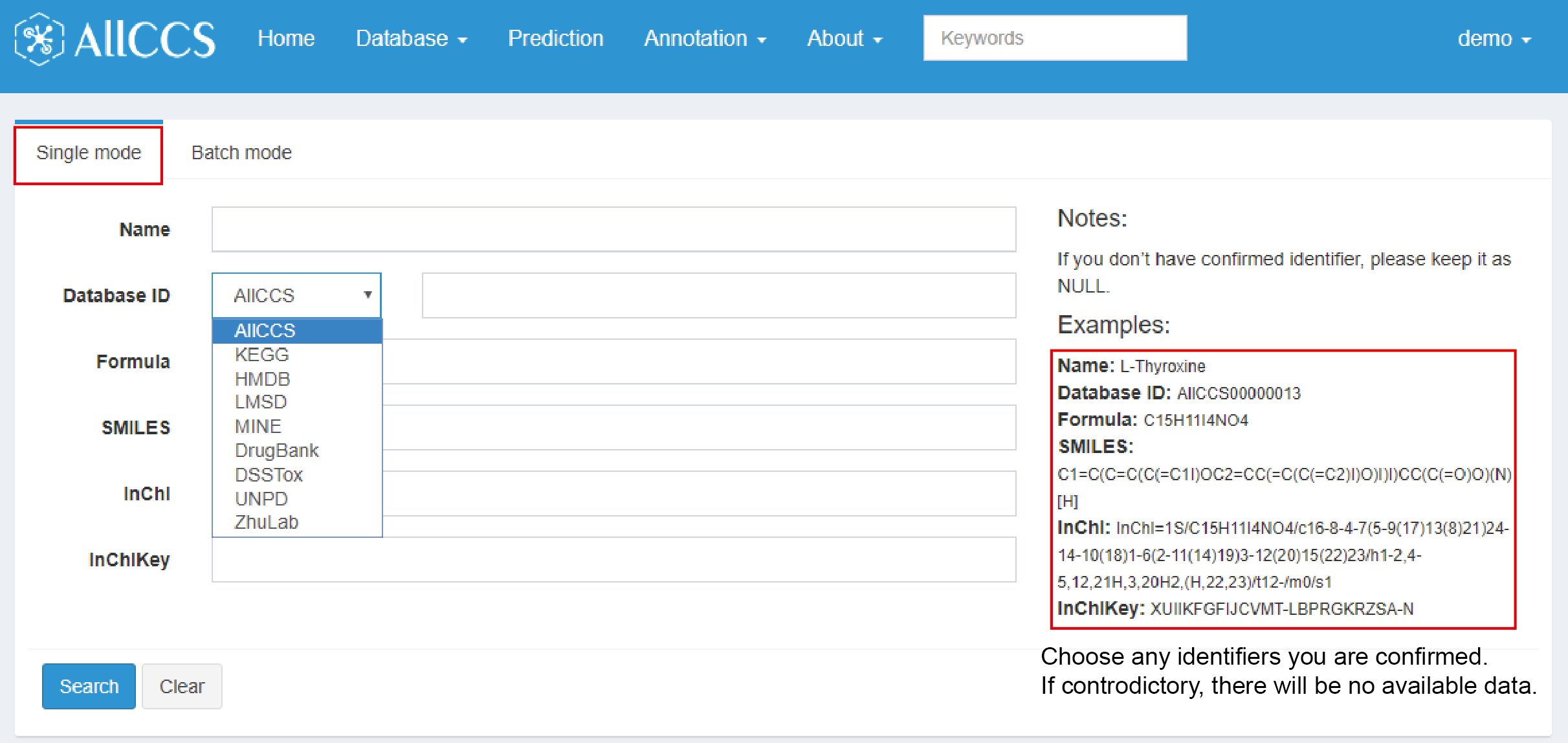
Figure 2.8: Advanced search – single mode
2.3.2 Batch mode
If there are multiple compounds, users can use batch mode to search (Figure 2.9). Currently, this function supports identifiers including “Database ID”, SMILES, InChIKey. While choosing database ID as identifier, there are several choice of databases in the right panel close to identifier option.
Note:
- In searching panel, you can enter one item per line.
- It should not contain extra space.
- It supports up to 100 query items per request.
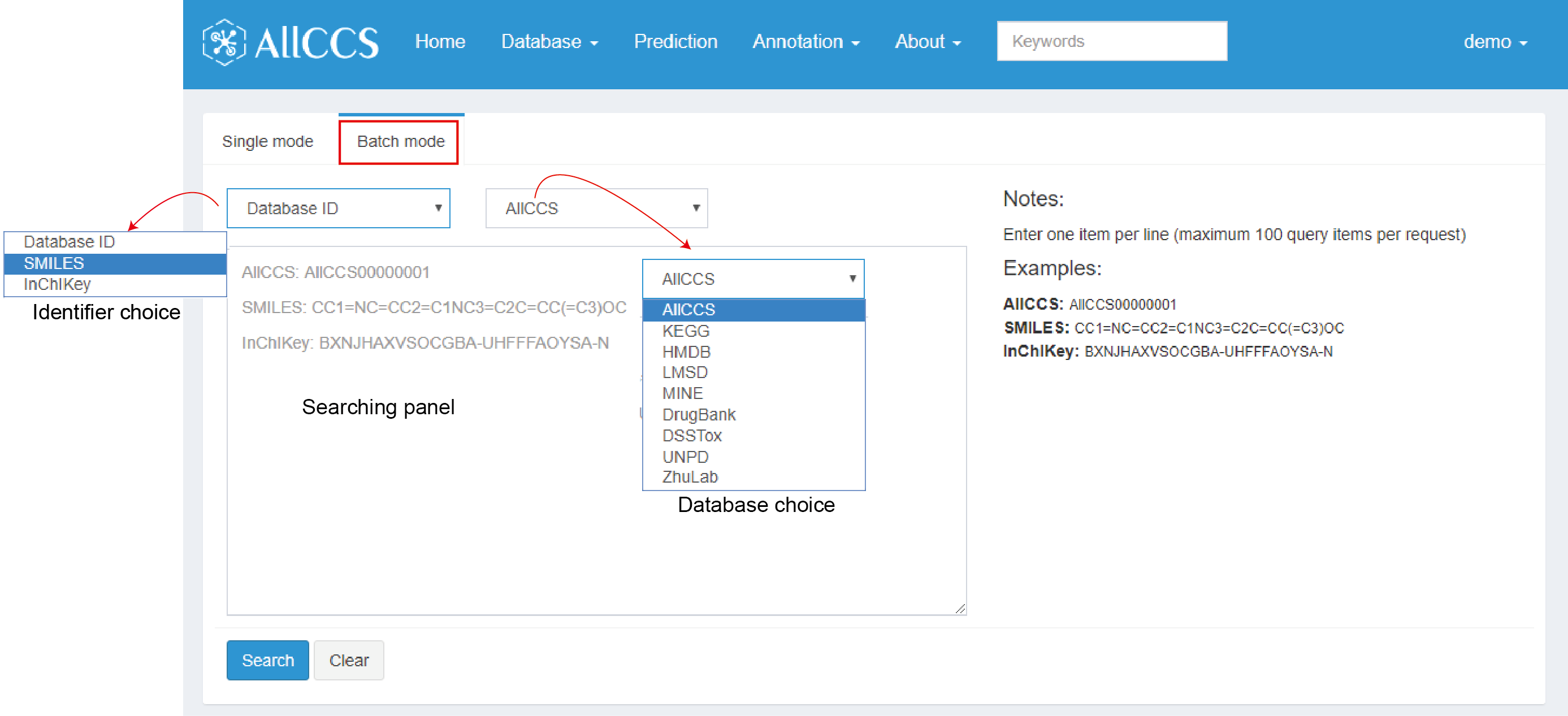
Figure 2.9: Advanced search – batch mode
References
Kanehisa, Minoru, and Susumu Goto. 2000. “KEGG: Kyoto Encyclopedia of Genes and Genomes.” Nucleic Acids Research, 27–30. doi:10.1093/nar/28.1.27.
Wishart, David S, Yannick Djoumbou Feunang, Ana Marcu, An Chi Guo, Kevin Liang, Rosa Vázquez-Fresno, Tanvir Sajed, et al. 2017b. “HMDB 4.0: The Human Metabolome Database for 2018.” Nucleic Acids Research, D608–D617. doi:10.1093/nar/gkx1089.
Fahy, Eoin, Shankar Subramaniam, Robert C. Murphy, Masahiro Nishijima, Christian R. H. Raetz, Takao Shimizu, Friedrich Spener, Gerrit van Meer, Michael J. O. Wakelam, and Edward A. Dennis. 2008. “Update of the Lipid Maps Comprehensive Classification System for Lipids.” The Journal of Lipid Research, S9–S14. doi:10.1194/jlr.R800095-JLR200.
Jeffryes, James G, Ricardo L Colastani, Mona Elbadawi-Sidhu, Tobias Kind, Thomas D Niehaus, Linda J Broadbelt, Andrew D Hanson, Oliver Fiehn, Keith E J Tyo, and Christopher S Henry. 2015. “MINEs: Open Access Databases of Computationally Predicted Enzyme Promiscuity Products for Untargeted Metabolomics.” Journal of Cheminformatics, no. 44. doi:10.1186/s13321-015-0087-1.
Wishart, David S, Yannick D Feunang, An C Guo, Elvis J Lo, Ana Marcu, Jason R Grant, Tanvir Sajed, et al. 2017a. “DrugBank 5.0: A Major Update to the Drugbank Database for 2018.” Nucleic Acids Research, D1074–D1082. doi:10.1093/nar/gkx1037.
M.Grulke, Christopher, Antony J.Williams, InthiranyThillanadarajah, and Ann M.Richard. 2019. “EPA’s Dsstox Database: History of Development of a Curated Chemistry Resource Supporting Computational Toxicology Research.” Computational Toxicology, no. 100096. doi:10.1016/j.comtox.2019.100096.
Gu, Jiangyong, Yuanshen Gui, Lirong Chen, Gu Yuan, Hui-Zhe Lu, and Xiaojie Xu. 2013. “Use of Natural Products as Chemical Library for Drug Discovery and Network Pharmacology.” Nucleic Acids Research, no. e62839. doi:10.1371/journal.pone.0062839.
Zhou, Zhiwei, Mingdu Luo, Xi Chen, and Zheng-Jiang Zhu. 2020. “Advancing Ccs Database Towards Metabolite Annotation.” In Preparing.
Feunang, Yannick Djoumbou, Roman Eisner, Craig Knox, Leonid Chepelev, Janna Hastings, Gareth Owen, Eoin Fahy, et al. 2016. “ClassyFire: Automated Chemical Classification with a Comprehensive, Computable Taxonomy.” Journal of Cheminformatics, no. 61. doi:10.1186/s13321-016-0174-y.